Which data can be shared through GBIF and what cannot
Preparing a dataset to be shared on GBIF.org can be a daunting task and many publishers realize that not all their data fits in the Darwin Core standard (DwC) and extensions GBIF uses to structure, standardize and display biodiversity data.
This blog post will cover what data fits in GBIF, give examples of data that does not fit in the current format of GBIF, and provide guidance to how you can share relevant data in a metadata-only dataset or through a third-party.
👍What data can you share on GBIF?
At its core, GBIF is a data infrastructure to share, view and retrieve global species occurrence data. All GBIF-mediated data comes from four different types of datasets published by endorsed organizations. GBIF users expect data to adhere to the four dataset types and are usually looking for species occurrences at a specific place and time. It is therefore recommended that publishers always keep the end users in mind when preparing their dataset for publication.
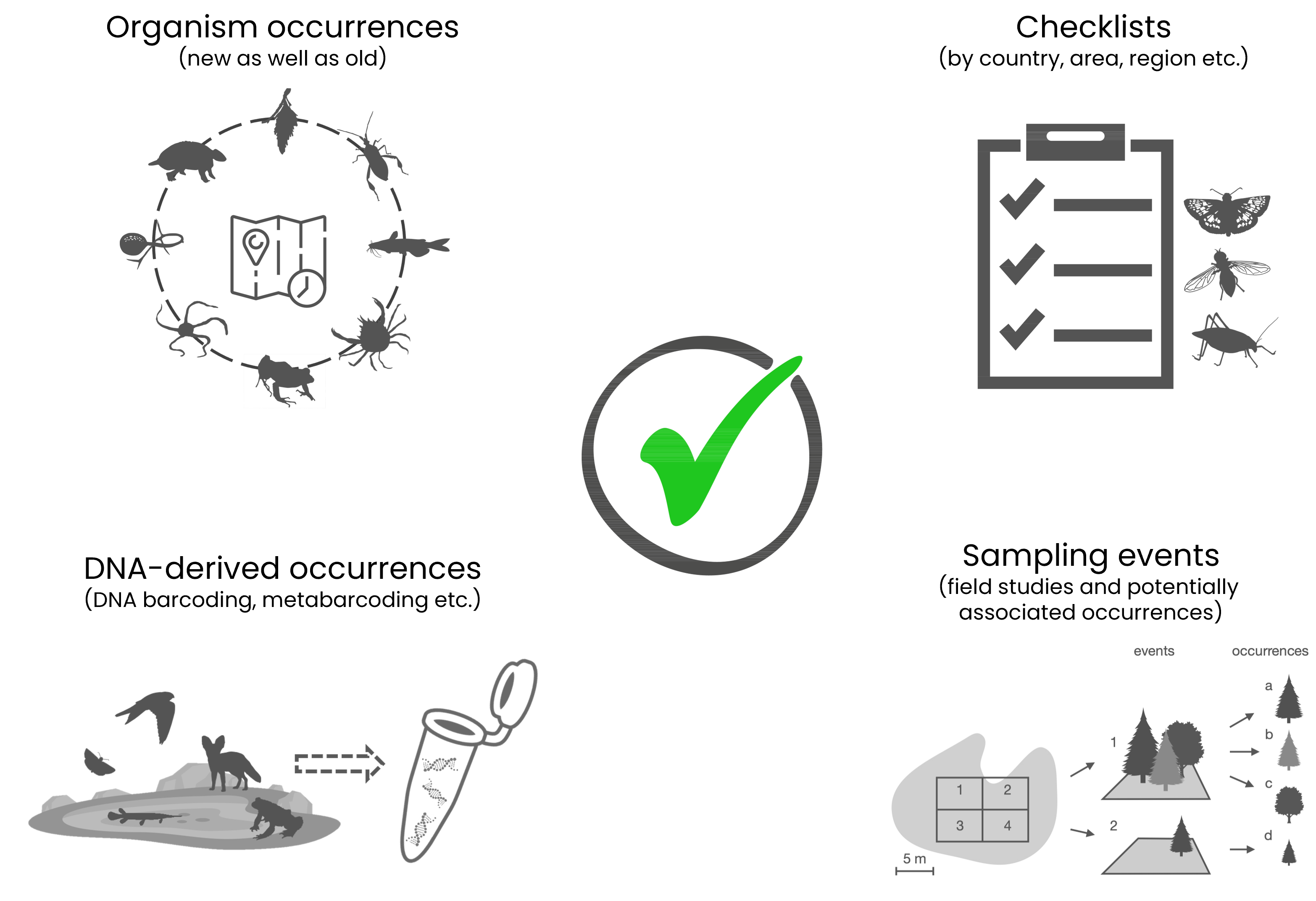
✔️Examples of data you can share in GBIF (the list is not exhaustive)
- the occurrence of a taxon at a given time and place (example, notice each term is a standardised DwC or extension field)
- DNA-derived occurrences (example, notice that this occurrence is in a cluster with other occurrences that represent the same organism in this occurrence and the occurrence record for the preserved specimen from which the DNA was obtained)
- species checklists for a country or area (example)
- sampling events (example of event record from an sampling event dataset, notice both present and absent occurrences were recorded)
- metadata-only datasets (example, no occurrences or events, just describing the data)
👎What you cannot or should not share on GBIF
Researchers, collections, monitoring professionals, projects, etc. often collect data that extends beyond species occurrences, which, to a large extent, can be captured in a sampling event dataset or in extensions. But what should you do if you have data that does not seem to fit in GBIF?
Data structure vs. data content
There are two main reasons why data is not shareable in GBIF:
- Data structure: the data structure is not supported within the DwC-Archive star schema GBIF uses to organize how data is related (the publication contains tables that relate to the extension files and not the core file, in other words - extension tables cannot have extensions themselves)
- Data content: the data has no associated DwC data standard field or you cannot find a suitable field in any of the extensions
Data structure
This blog post focuses on the data content (reason #2), but if you want to familiarize yourself with the schema and the associated data sharing limitations, you can read more in the Darwin Core Archive how-to-guide.
The central idea of an archive is that its data files are logically arranged in a star-like manner, with one core data file surrounded by any number of > ‘extension’ data files. Core and extension files contain data records, one per line. Each extension record (or ‘extension file row’) points to a record in > the core file; in this way, many extension records can exist for each single core record. This is sometimes referred to as a “star schema”.
GBIF is aware that the star-shema structure of the archives are limiting to publishers and we are working on a solution to overcome such data structure limitations. If you want to follow the work, please take a look at the new data model work.
Data content
GBIF has seen some creative ways to make the DwC standard fit to other types of data. The creativity tends to arise when researchers have (super cool!) complex, data-rich research projects often involving novel observation methods, or when scientific collections want to digitize additional information from their collections.
Once we had to delete a dataset since all the occurrences were in the future. The project modelled probable occurrences to help inform policymakers of likely outbreaks of wheat disease, but GBIF does not support potential future occurrences.

❌Examples of data that does not fit in GBIF (the list is not exhaustive)
- observations of humans (although they occasionally make their way into citizen science and collections datasets)
- datasets based on data extracted from GBIF and somehow modified or cleaned
- species occurrences in extraction blanks and PCR negatives from DNA-derived occurrence datasets
- future occurrences (like the example above)
- occurrences of non-organisms (for example ecosystems, sampling areas, national parks, rocks (although we do support fossil specimen occurrences))
How to use metadata-only datasets to make your work more visible
Metadata-only datasets can be used to share information of data not available online, including data that does not fit in the other dataset classes and the associated extensions.
For example, collections can use metadata-only datasets to describe undigitized resources in their collections. Metadata-only datasets also receive a DOI upon publication similarily to the other dataset classes, so information in metadata datsets can be cited, making the information shared in the dataset more discoverable and reusable.
Examples of metadata-only datasets in GBIF.org
A metadata-only dataset might contain information on external data resources not published in GBIF, but findable in other places. For example, to share that your organization provided data for the 2nd European Breeding Bird Atlas or has a database on the biodiversity of the Altai-Sayan ecoregion.
Another metadata-only dataset could share that a survey, sampling event or project was carried out, even though the data is not shared with GBIF, e.g. a plankton survey the area of the Northern Antarctic Peninsula.
Complimentary to metadata-only datasets in GBIF, external repositories, e.g. Zenodo, can be used to share data, code, etc.
Sampling and lab protocols and bioinformatics pipelines, can be shared on protocols.io which allows for a more stuctured and reusable sharing of protocol steps compared to what can be captured in GBIFs metadata section. Both Zenodo and protocols.io issues DOIs when datasets are published which can be referenced in the GBIF metadata-only dataset to increase discoverability.
💡Bonus: Example of how to share data from a sampling event - and how not to share the data
This section does not strictly cover data that does not fit in a GBIF sampling event, occurrence or checklist dataset. Rather, it covers an example of data from multispecies surveys that do fit into GBIFs publishing model, but where new publishers may be unsure how best to structure their dataset(s).
Example
An entomologist carries out sweep-net sampling of five different areas. Sweep-net sampling is carried out walking a transect, sweeping 10 times along the way, which yields a sample of multiple species each time.
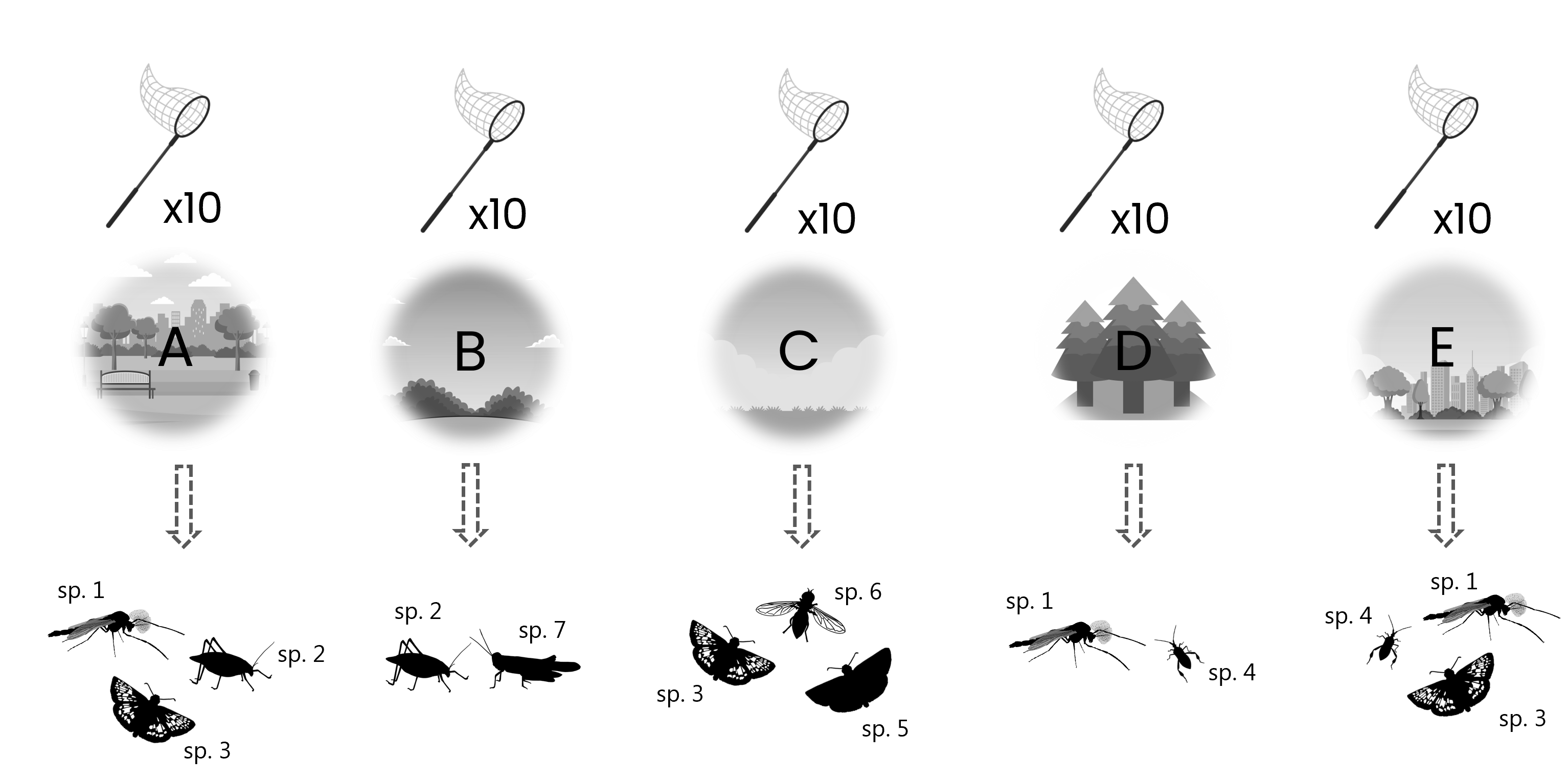
This type of data is best captured in an event table and an occurrences table, where the sampling event data goes in the event table and the species occurrences go in occurrence table, similar to the example previously.
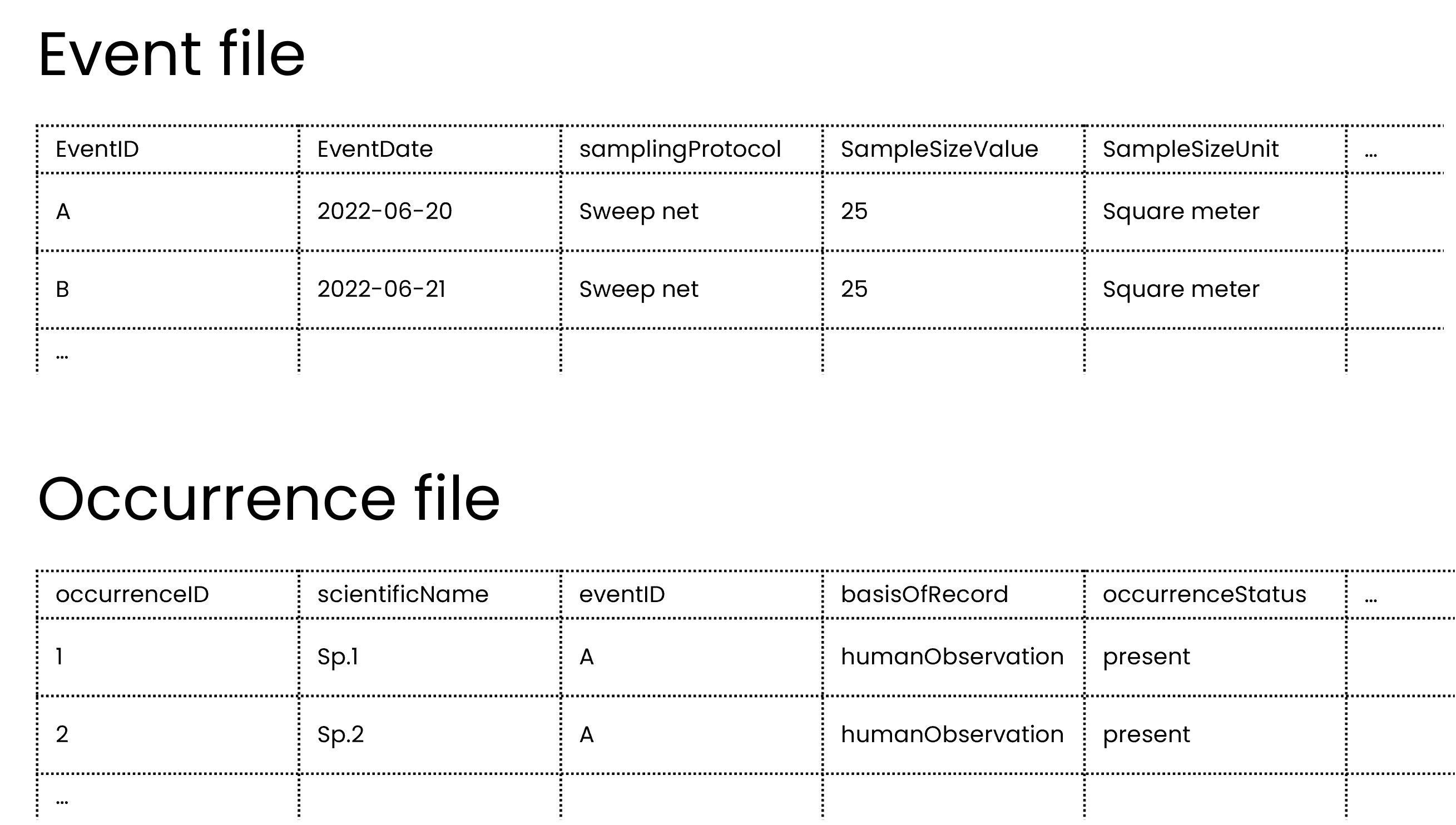
As mentioned in previously, publishers should not share occurrences of non-organisms so in this case, the publisher should not share the multi-taxa sampling event as an occurrence.
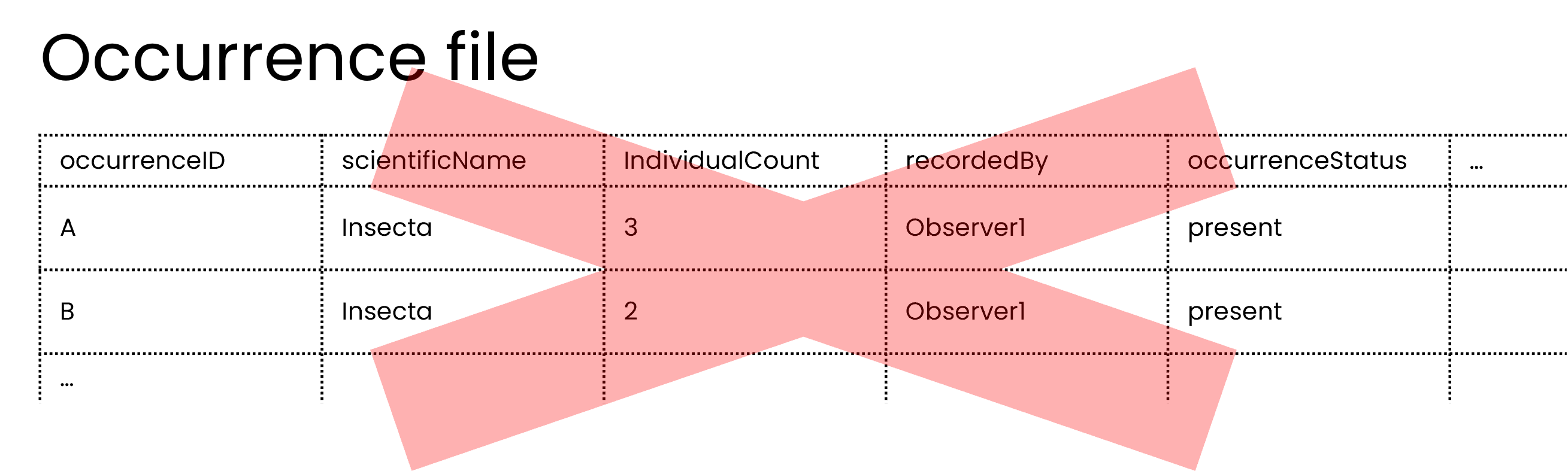
Technically, the occurrence file could be shared, but if useability is considered, then the example with an event file and an occurrence file yields much richer data and allows users to find specific taxa and discern between the sampling event and the occurrences.
You can read more on how to choose dataset types in this blogpost.
NOTE! if you are in doubt if and how your data fits into GBIF, you can always contact helpdesk@gbif.org and ask for guidance.
