Identifying potentially related records - How does the GBIF data-clustering feature work?
NOTE: The content of this blog post is now maintained in the technical documentation. Please view this source for the most up to date information on the topic.
Many data users may suspect they’ve discovered duplicated records in the GBIF index. You download data from GBIF, analyze them and realize that some records have the same date, scientific name, catalogue number and location but come from two different publishers or have slightly different attributes.
There are many valid reasons why these duplicates appear on GBIF. Sometimes an observation was recorded in two different systems, sometimes several records correspond to herbaria duplicates (you can check the work of Nicky Nicolson on the topic), sometimes a specimen was digitized twice, sometimes a record has been enriched with genetic information and republished via a different platform…
This is why last year, we released an experimental data-clustering feature aiming to identify potentially related records on GBIF. The goal was not only to detect potential duplicates but to also find interesting relationships, such as between typification records or records originating from Natural History collections, DNA-derived sequences and citations of materials examined when publishing taxonomic treatments in journal articles.
Records that have been included in a cluster can be found with the “is in cluster” filter in the occurrence search. Each occurrence page belonging to a cluster will have a “CLUSTER” tab displaying the potentially related records (see a screenshot of this example below).
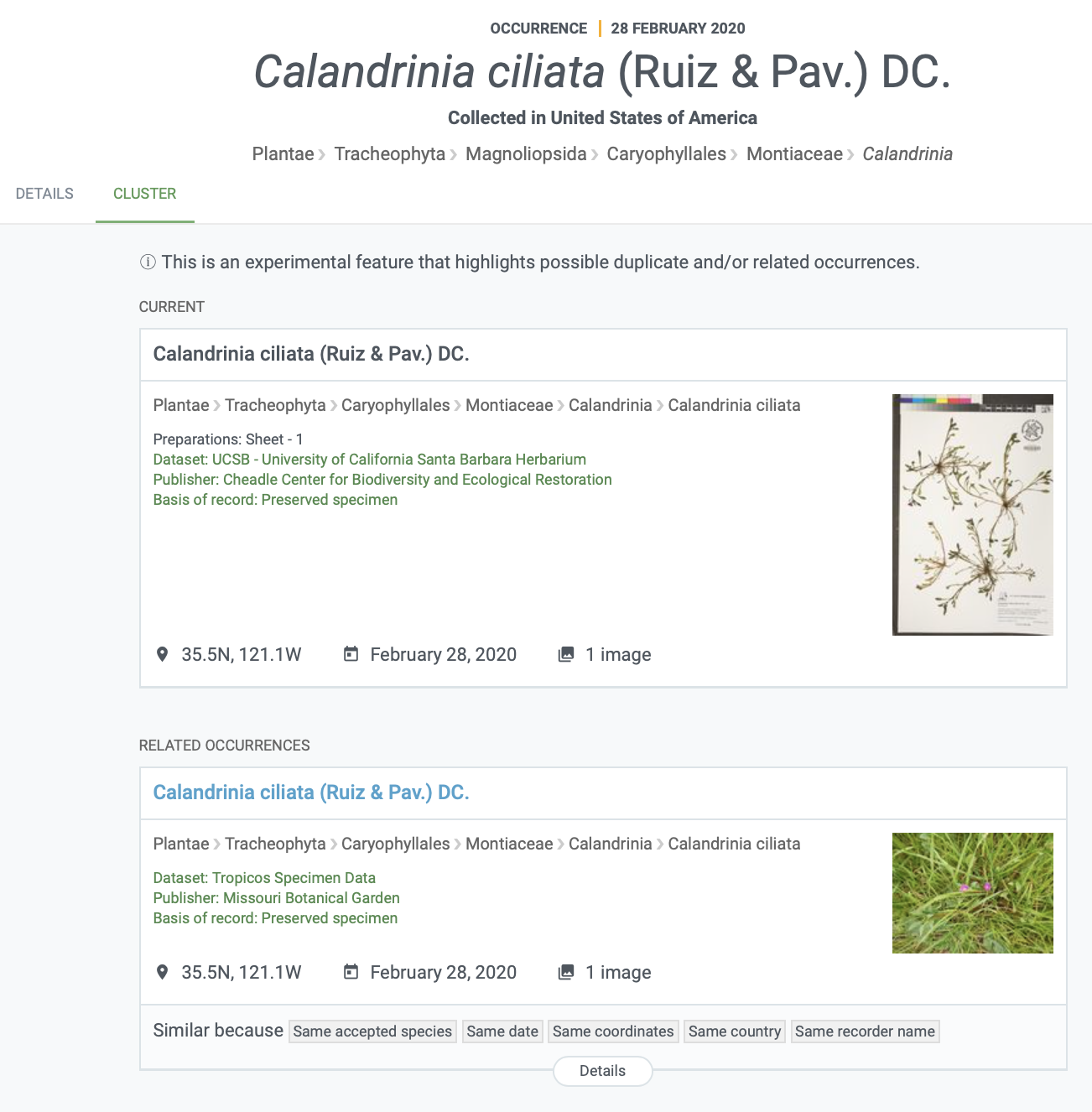
You can read this news item for more information and some exciting examples.
How does the GBIF data-clustering feature work?
Step 1: Select candidates
Comparing nearly 2 billions records with each other is very resource intensive and quite impractical, so the first step of the data-clustering process is to select and group candidate records to compare.
The system first creates a series of “hashes” for each record based on specified fields. All records sharing a hash are candidates to compare against each other.
For example, one of the “hashes” used is based on the species key, rounded coordinates, year, month and day. This means that the records that share the same values for those fields will be grouped together in the candidate table for further inspection.
The fields used to identify and group the candidates are a subset of what is used later on for comparing them (see the table below). See this link to check the details.
Step 2: Compare candidates with each other and assess whether they are potentially related
In this second phase, the system will compare all records in the candidate set to each other and generate assertions. The assertions are inspected and the algorithms decides if there is enough evidence in the assertions to create a relationship between them. In the table below, I summarize how those assertions are made but if you would like more details, you can check the code available here.
[Edit 2022-06-23 : the table below has updated to reflect some changes in the clustering function https://github.com/gbif/pipelines/issues/733]
| Assertion | fields checked | condition checked |
|---|---|---|
| Same specimen | taxonKey, typeSatus |
same taxonKey between records and type status = Holotype for both records |
| Typification relation | scientificName, typeStatus |
same between records |
| Same accepted species | speciesKey |
same between records |
| Same date | eventDate or day, month, year |
same between records |
| Approximate date | day, month, year |
dates one day apart |
| Different date | eventDate |
differs between records |
| Non conflicting date | eventDate, day, month, year |
no date on either record |
| Same recorder name | recordedBy |
same between records |
| Same coordinates | decimalLatitude, decimalLongitude |
same between records |
| Non conflicting Coordinates | decimalLatitude, decimalLongitude |
no coordinate on one or both sides |
| Within 200 m | decimalLatitude, decimalLongitude |
distance <= 0.200 |
| Within 2 km | decimalLatitude, decimalLongitude |
distance <= 2.00 |
| Same country | countryCode |
same between records |
| Non conflicting country | countryCode |
country only on one record |
| Different country | countryCode |
differs between records |
| Identifiers overlap | occurrenceID, fieldNumber, recordNumber, otherCatalogueNumber, grouped(institutionCode:collectionCode:catalogueNumber),grouped( institutionCode:catalogueNumber) |
checks any overlap of identifiers between records |
| Other catalogue number overlap | otherCatalogueNumber,grouped(institutionCode, collectionCode, catalogueNumber) with any separator checks if the other catalogue number correspond to the institution code, collection code and catalogue number from another record |
|
| Is from sequence repository | datasetKey |
checks if one of the datasetKey corresponds to one of the sequence repository datasets: INSDC sequences, INSDC host organisms, INSDC environmental samples, iBOL (see keys d8cd16ba-bb74-4420-821e-083f2bac17c2, 393b8c26-e4e0-4dd0-a218-93fc074ebf4e, 583d91fe-bbc0-4b4a-afe1-801f88263016 and 040c5662-da76-4782-a48e-cdea1892d14c) |
| Are specimens | basisOfRecord |
checks if the basis of record for both records are one of the following: PRESERVED_SPECIMEN, LIVING_SPECIMEN, FOSSIL_SPECIMEN, MATERIAL_CITATION |
The table below summarises the combinations of assertions that are sufficient to link the records in a cluster. If a group of occurrences share the combinations of assertions for any given column, they will be clustered together. [Edit 2022-03-11: The table below has been updated to reflect some changes in the clustering function: https://www.gbif.org/release-notes#11mar22] [Edit 2022-06-23 : the table below has updated to reflect some changes in the clustering function https://github.com/gbif/pipelines/issues/733]
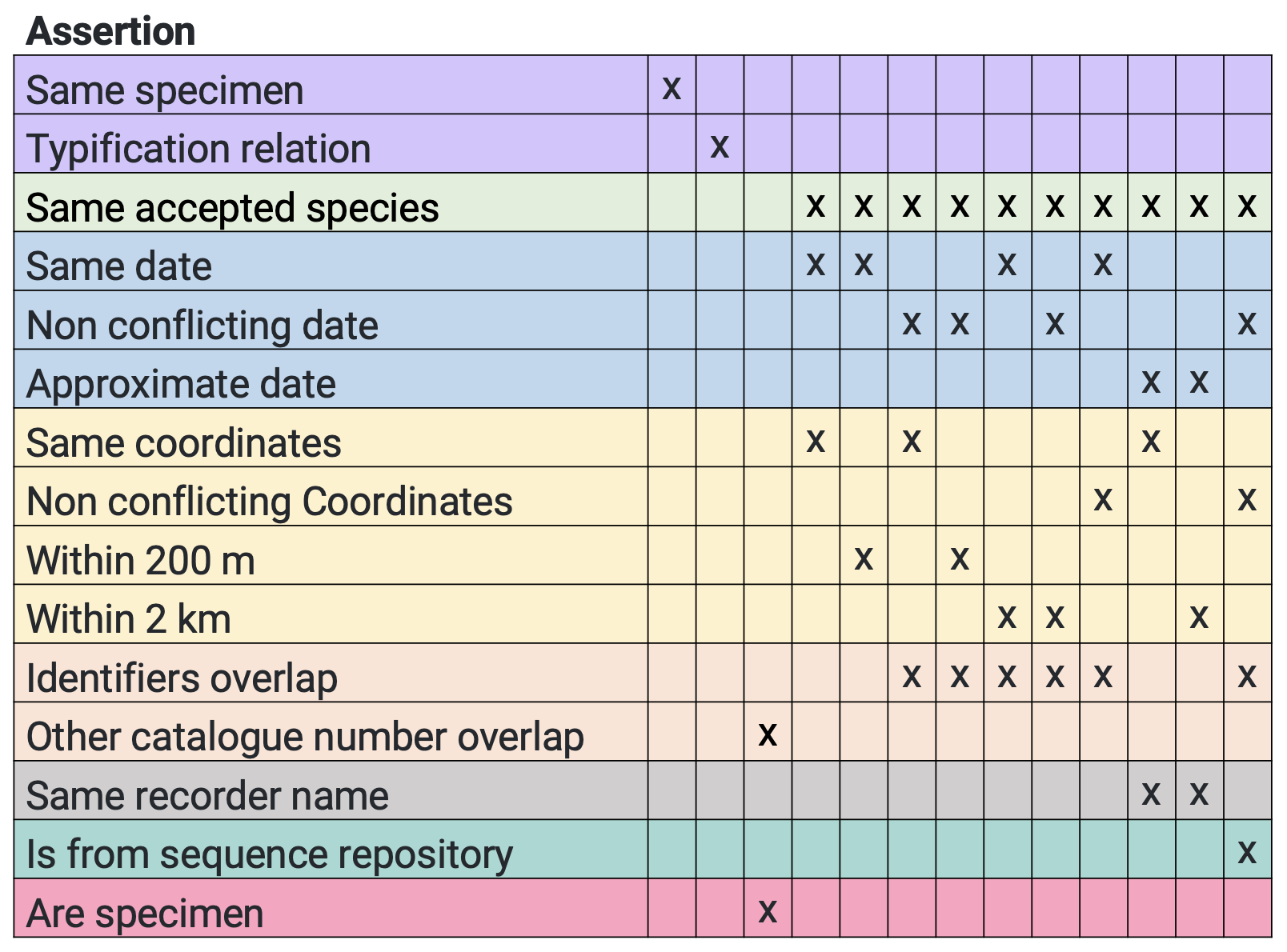
NOTE: Any group of occurrences associated with the assertion
Different dateorDifferent countrywill not be clustered together.
Why some occurrences are not clustered
It is possible that some occurrences check one of the combinations of assertions but aren’t shown as clustered yet. This could be the case for several reasons:
- The occurrences are newly published. Right now, the clustering process is quite resource intensive and doesn’t run automatically. We need to trigger it manually. This means that it can take a few weeks before newly published occurrences get clustered.
- The “duplicates” come from the same dataset. The clustering algorithm only compares occurrences across datasets, not within datasets.
- There can be a delay between the moment the occurrences are clustered and the moment they become searchable with the “is in cluster” filter (this is due to some technical reasons a bit too long to explain in this post, but relate to updating the search indexes separately from the clustering table)
There could be other unforeseen reasons, and if in doubt, please contact us at helpdesk@gbif.org.
What can you do to improve linkage with other occurrences?
If for one reason or another, you need to publish on GBIF occurrences for observations or specimens that you know are already on GBIF, how best to do it?
- Make sure that you reuse the same identifiers as much as possible, including the formating. Same catalogue numbers, occurrenceID, etc.
- Use the associatedOccurrences and resource relationship extension. These are not used during the clustering today, but are expected to be in the future, and are the correct way to communicate relationships within Darwin Core.
Where to send suggestions and ask questions
If you have suggestions to improve the clustering feature or questions on how it works, you are very welcome to:
- leave a comment under this post
- or log an issue in our GitHub repository or via the GBIF feedback system
- or contact us at helpdesk@gbif.org.
