[Edit 2021-09-16]
Important: To find guidance on how to publish Sequence-based data on GBIF, please consult the following guide:
Andersson AF, Bissett A, Finstad AG, Fossøy F, Grosjean M, Hope M, Jeppesen TS, Kõljalg U, Lundin D, Nilsson RN, Prager M, Svenningsen C & Schigel D (2020) Publishing DNA-derived data through biodiversity data platforms. v1.0 Copenhagen: GBIF Secretariat. https://doi.org/10.35035/doc-vf1a-nr22.
[End edit 2021-09-16]
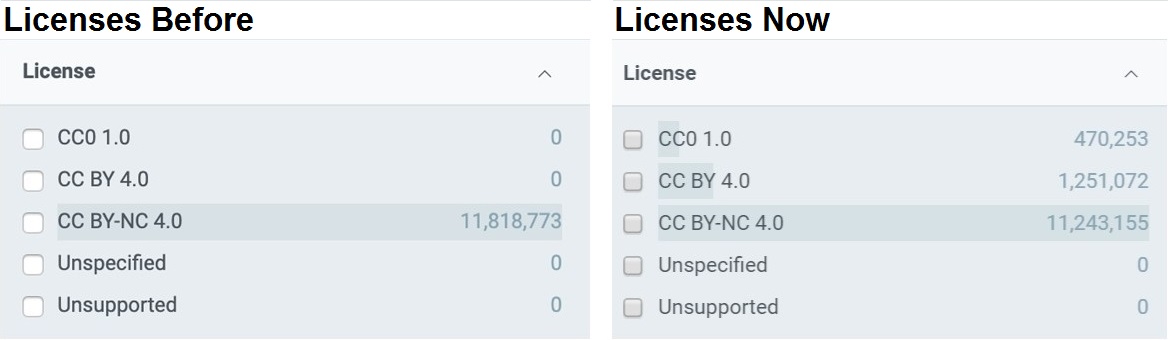
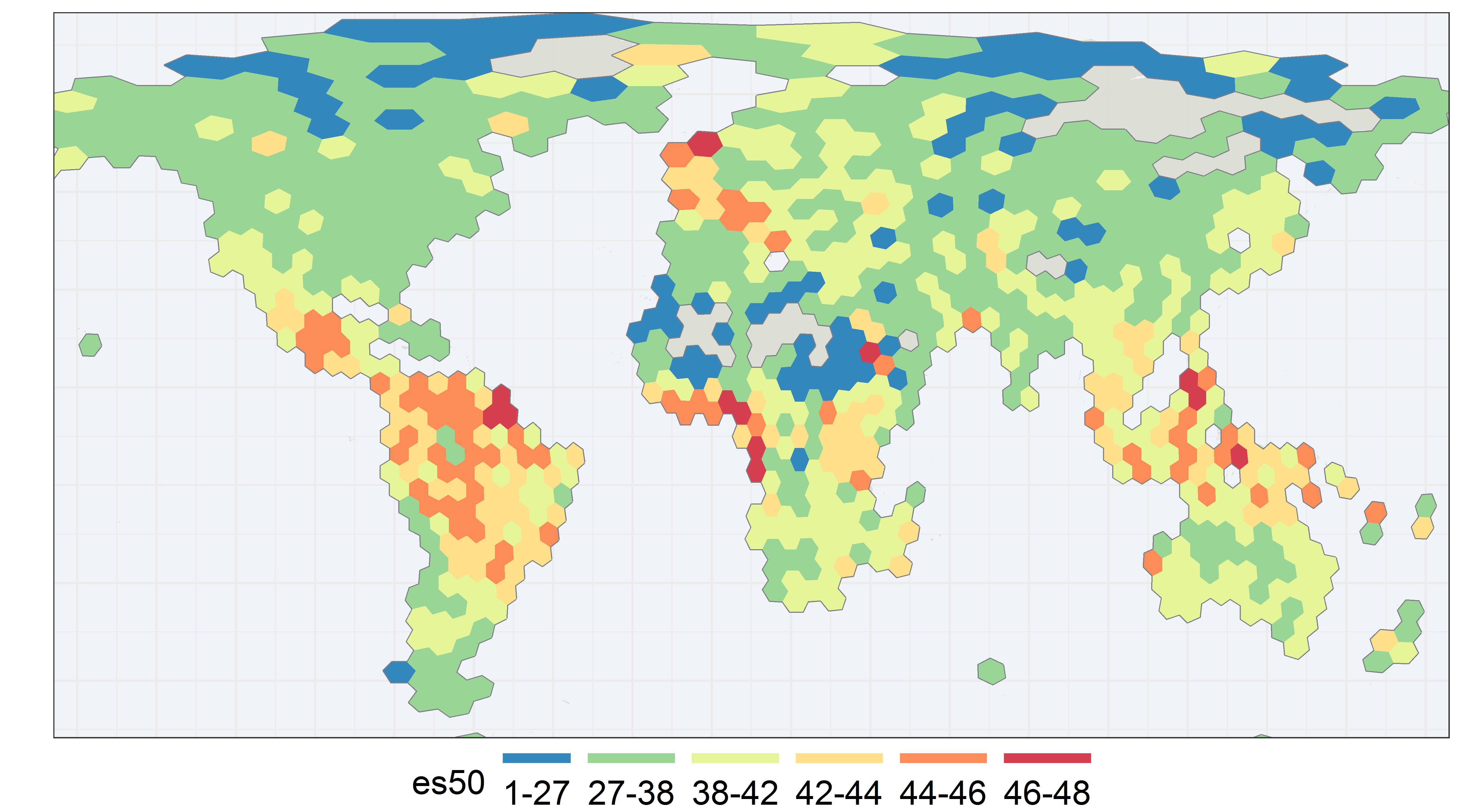
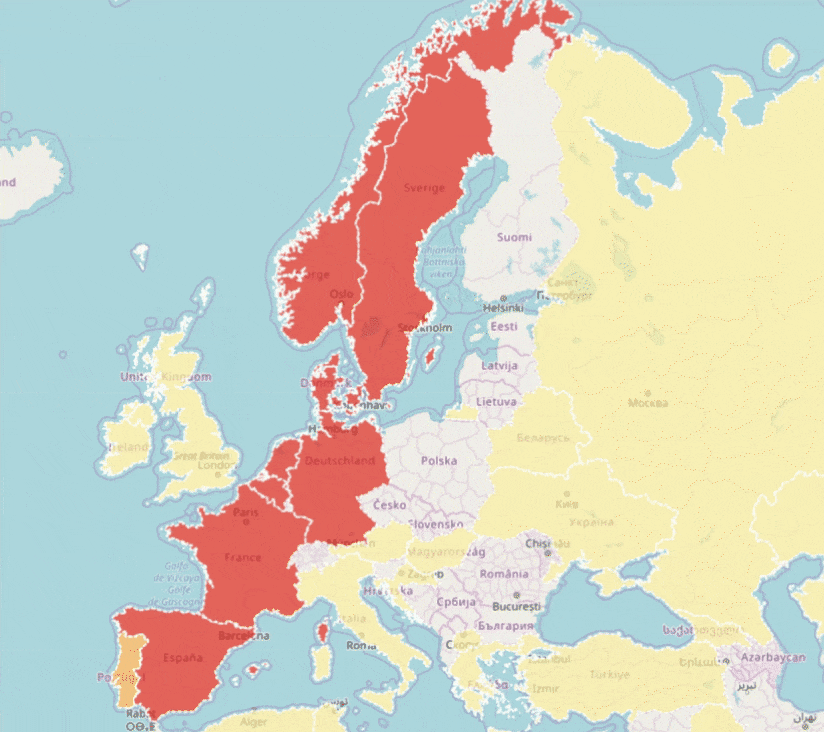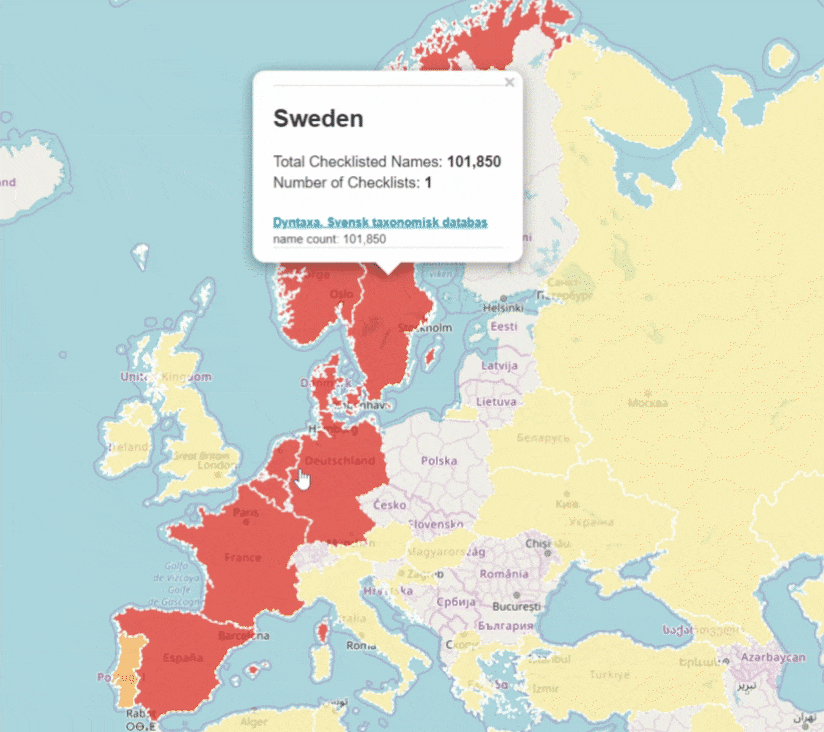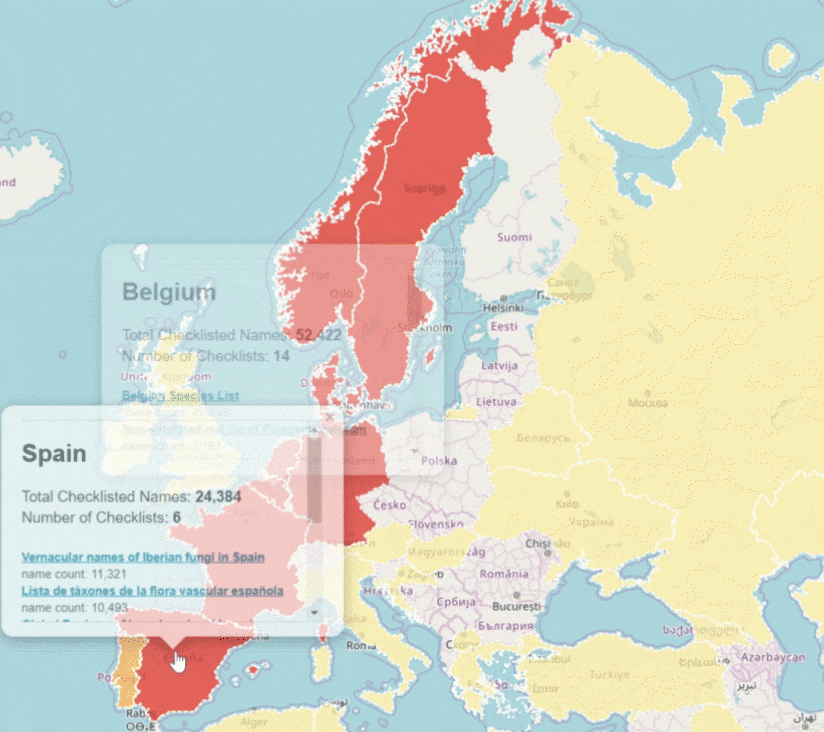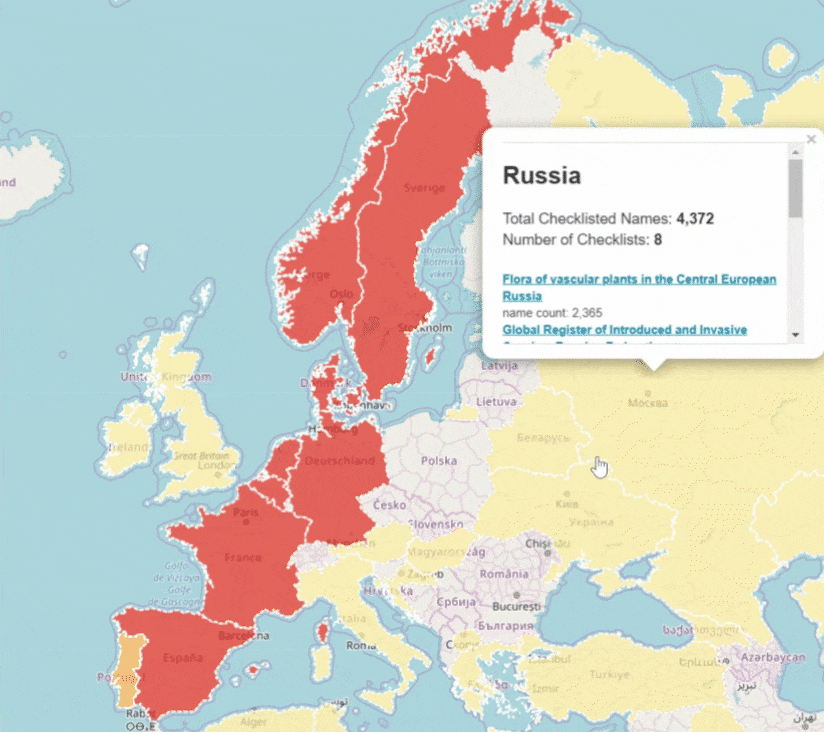
GBIF is trying to make it easier to share sequence-based data. In fact, this past year alone, we worked with UNITE to integrate species hypothesis for fungi and with EMBL-EBI to publish 295 metagenomics datasets.
Unfortunately, documentation is not as quick to follow. Although we have now an FAQ on the topic, I thought that anyone could use a blog post with some advice and examples.
Note that this blog post is not intended to be documentation. The information written is subject to change, feel free to leave comments if you have any question or suggestion.