(Almost) everything you want to know about the GBIF Species API
Today, we are talking about the GBIF Species API. Although you might not use it directly, you probably encountered it while using the GBIF web portal:
- Typing a scientific name in the GBIF Occurrence search.
- Seeing a “Taxon Match Fuzzy” flag.
- Using the Species Name matching tool.
This API is what allow us to navigate through the names available on GBIF. I will try to avoid repeating what you can already find in its documentation. Instead, I will attempt to give an overview and answer some questions that we received in the past.
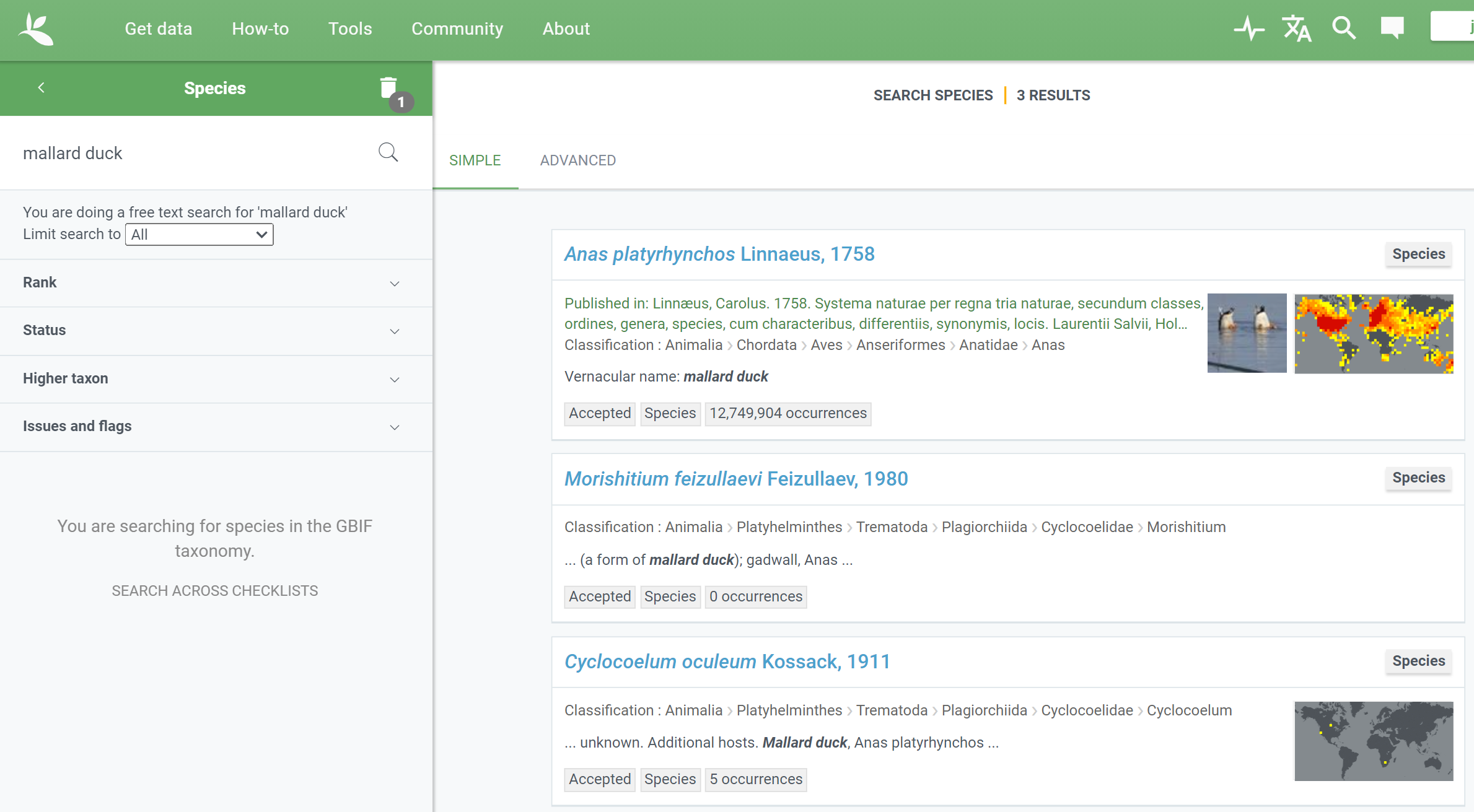
What can the species API do and where is it used?
If you head over to the API documentation page, you will see that it divides the functions in three categories:
- Working with Name Usages: these are all the calls use to navigate the GBIF backbone taxonomy or any other checklists available on GBIF. They are used by the species web interface. More information here.
- Searching Names: these are four functions to search for names.
- The
/species/searchfunction is used to query the taxon names on the species web interface. - The
/species/suggestfunction is used by the drop down menu that appear when searching names in theScientific namefield in the occurrence search interface. - The
/species/matchfunction is used to match names to the GBIF backbone taxonomy. When occurrences are shared on GBIF, all the scientific names are matched to the backbone taxonomy using this function. The Species Name matching tool also uses this function. Note that for names included in checklists, an other function, more stringent, is used. We assume that names included in checklists are carefully curated and don’t need fuzzy matching (as this is the goal of a checklist). - The
/speciesfunction is not used in the GBIF Portal but it allows to search for exact matches in any checklist you would like.
- The
- Name Parser: these are the calls used by the
Searching Namesfunctions as well as GBIF Name Parser tool.
What does it mean when some fields are not included in the response?
This means that we don’t have the value for that particular taxon and field.
Search and Match
Both the search and match functions are based on the Lucene search technology. We use an analyzer which takes into account specificities of text matching for scientific names. However, there are a few key differences between the two functions.
The search function queries everything (name, description, etc.) and the result is ranked according to where the match was found. See the figure below:
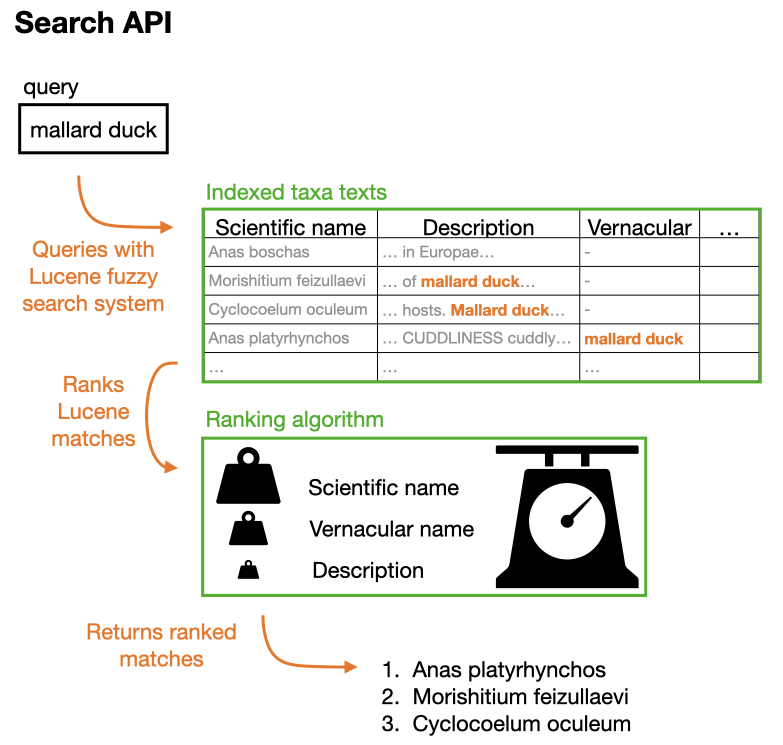
Note that it is possible to specify the field searched thanks to the qField parameter. For example, if you would like the algorithm to check vernacular names specifically, you can write qField=VERNACULAR.
The code corresponding to the Search function can be found here.
The match function uses the fuzzy matching on canonical names to generate candidate matches. The scientific authorships are excluded for the first step of the matching because they vary so much. The resulting matches are then scored according to their taxonomy, authorship, status and rank (the scores are available in the note field of the API response by setting the parameter verbose=true).
For a summary of the match function, see the figure below:
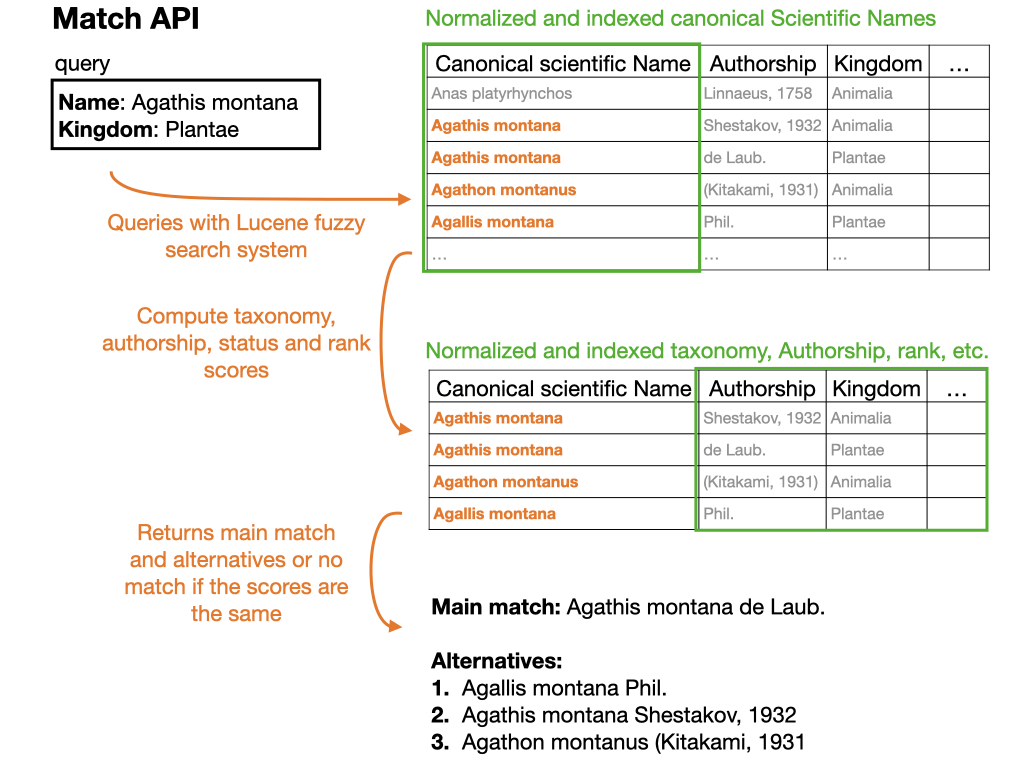
The example used in the figure is the following:
https://api.gbif.org/v1/species/match?verbose=true&kingdom=Plantae&name=Agathis%20montana
The matching algorithm generates a number of flags. The main one being “Taxon Match Fuzzy”, which means that the match found doesn’t exactly match the query (different spelling). More information about GBIF flags and issues in this blogpost.
If two matches score the same, nothing is returned as main match but they can be found as alternative matches (which are available in the response when setting the parameter verbose=true). This is why providing a higher taxonomy along with your scientific name can really improve the matching: it will weigh on the taxonomic score and help find a “main match”.
Note that not all taxonomic ranks are created equal in our scoring system. Because there can be so much variation in the taxonomies available, a difference at the order level doesn’t weigh as much a as a difference at the kingdom level. In addition to that, our algorithm is more stringent when it comes to differentiate Animalia from Plantae than other kingdoms (Protista vs Animalia or Chromista vs Plantae, etc.)
The code corresponding to the match function can be found here.
Is it possible to get the result of a species search as an excel/CSV table?
No, there is no “species download” like we have for occurrences. You will have to use the API and convert the JSON result into a CSV/excel table. There are a number of online JSON converters you can try if you are not comfortable with writing your own scripts.
For more information
If you would like to know more about the GBIF backbone taxonomy and how it is generated, you can check out this blogpost. You are also very welcome to send your questions and report any issue via our feedback system or on GitHub here.
Let me know if this post is useful and if there is anything I left out that you would like to know more of. Thanks!
