Making collection content discoverable when you don’t have occurrences published on GBIF
This blog post is a tutorial on how to upload collection descriptors in the Global Registry of Scientific Collections (GRSciColl).
If you are someone working with a physical collection or work with people who do, you might be interested in making these collections findable. Ideally, the content of these collections would be digitized and made available online on relevant platforms like GBIF.org, iDigBio.org, ALA.org.au, etc. Sharing digital specimen records is a great way to ensure the discoverability of collection content. Not only users can find your collection but can also use individual records relevant to their research. If you can publish occurrences on GBIF, please do it!
However, not everyone can share records at the specimen level on GBIF. For example, you might be working with a mineralogy collection where the data would be outside of the GBIF scope. Or perhaps you only have partial information about your collection: for example, you might only have the number of jars of amphibians or fish.
If that’s your case, you might be interested in packaging the data you have as collection descriptors and uploading them to the Global Registry of Scientific Collections (GRSciColl).
A brief reminder of what GRScicoll is
The Global Registry of Scientific Collections (GRSciColl) is a registry of physical scientific collections (content, location and contacts) and associated institution. GRSciColl has its own history and scope. It was originally developed and maintained at the Smithsonian Institution before GBIF inherited it. You can read more about how here and here.
GRSciColl and GBIF have overlapping content: many of the collections listed in GRSciColl are also datasets on GBIF. However, the scope of GRSciColl goes beyond biodiversity data, for example, GRSciColl contains mineralogy and archaeology collections but doesn’t include things like citizen science observation data.
You can find out more about GRSciColl and browse the GRSciColl data here: https://scientific-collections.gbif.org. You can also watch this short introduction below.
What are collection descriptors in GRSciColl?
Collection descriptors were implemented as part of the GRSciColl 2024 roadmap. They are meant to share structured information about collections. They can contain relevant details about collections and sub-collections as well as quantitative data which cannot be shared on collection pages (for example, the number of type specimens for a particular taxon). Some collection descriptors are used for indexing collections. This means that they improve collection discoverability. For example, a collection entry associated with dragonfly species names will be found by users looking for “Odonata” in the scientific name field of the collection search.
Here are some examples of searches based on collection descriptors:
- Find some collections with specimens collected by M. J. Berkeley
- Find some fern collections hosted in French institutions
Currently, only a handful of collection descriptors are indexed and searchable: scientific name, country or area of coverage (of the specimen, this is based on the dwc:country term), recorded by and type status. However, GBIF will be able to add filters for more standardized terms as more descriptors are shared in GRSciColl.
When to upload collection descriptors in GRSciColl:
- The collection is not part of the GBIF Scope (for example, mineralogy, archeology, anthropology, etc.)
- The information about the collection is structured but not at the specimen level (e.g. you can not create occurrences).
- You have information that does not fit the Darwin Core Standard.
How to format collection descriptors
Each GRSciColl collection entry can have one or several collection descriptor groups. A group can correspond to descriptors for a particular aspect of the collection or a sub-collection. Each group requires:
- A title of the set of descriptors. For example, “Taxonomic breakdown of the algae sub-collection”.
- A description for the set of descriptors. For example, “These descriptors are based on the 2008 inventory of the algarium. This inventory focused mainly on type specimens”.
- A comma-separated file containing the descriptors where each column is a descriptor and each row a subset of the collection described. The header of the table is used to map its content to Darwin Core and Latimer Core (see more details below).
The descriptor table
As mentioned above the descriptor tables are CSV files where each row is a subset of the collection (or a group of specimens) and each column is a descriptor.
When possible, the data should be mapped to the Darwin Core and Latimer Core standards but it is possible to share data that is not mapped to any standard. When mapped to one of the standards, the header of the column should contain the prefix of the standard (ltc: for Latimer Core and dwc: for Darwin Core) as well as the name of the term.
A table could look like this fictional example:
| ltc:biomeType | dwc:scientificName | dwc:country | Number of identified specimens at genus level |
|---|---|---|---|
| Freshwater | Perciformes | Colombia | 300 |
| Freshwater | Perciformes | Brazil | 145 |
In the example above, the last column couldn’t be mapped to any Darwin Core or Latimer Core term so it was left with a descriptive title. This column will not be indexed, and users will not be able to search data based on its values, but it will be displayed along the other descriptors on the collection page. See an example of collection descriptors for The New York Botanical Garden collection where not every column is mapped to a standard:
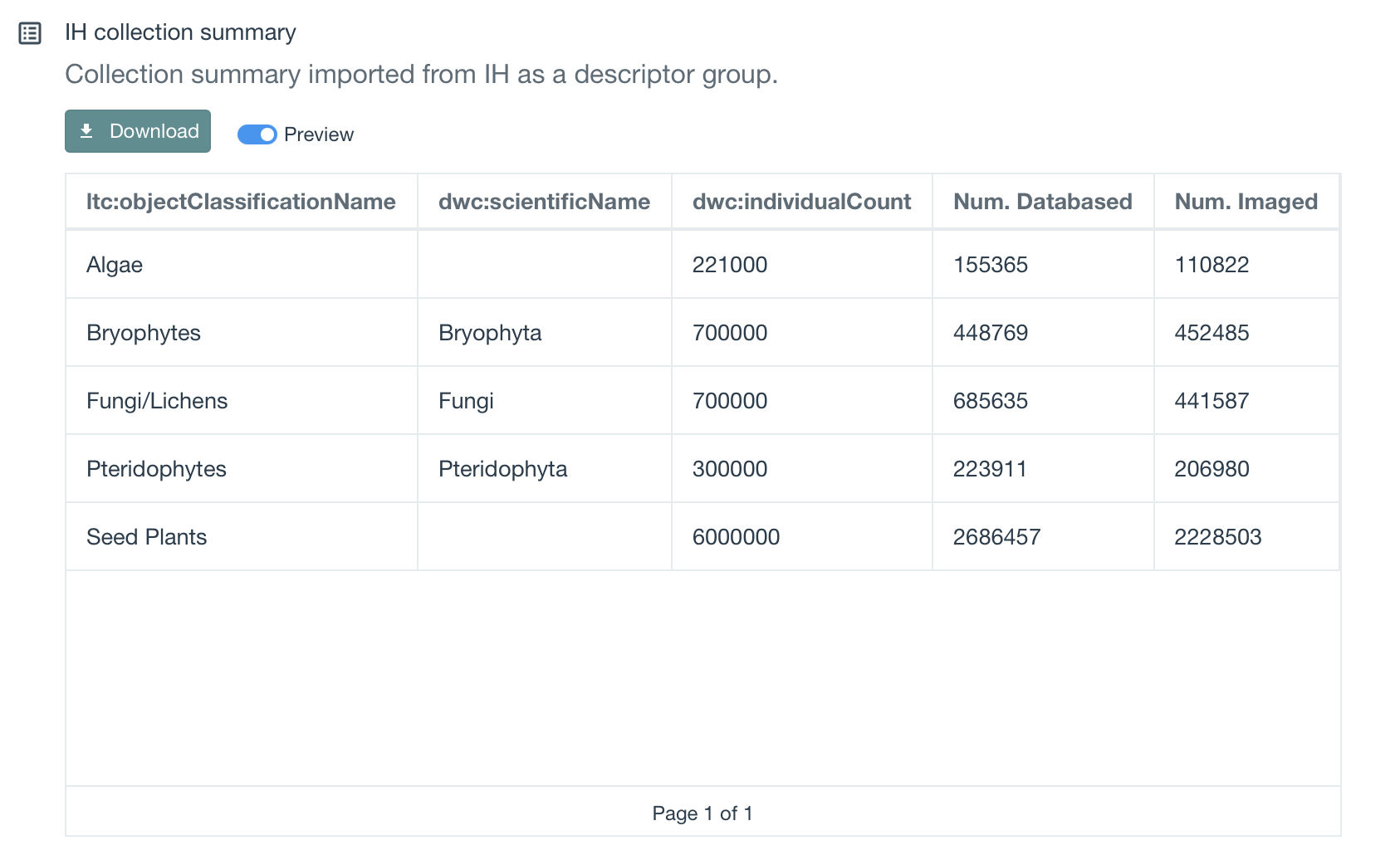
Note that tables might contain overlapping information or different descriptions for the same subset of specimens.
There is not any template to download as the descriptors can include a lot of headers. You are very welcome to download any table you like from GRSciColl and use it as your own template. Here are some examples that I compiled from real data while working on the implementation of descriptors. They could certainly be mapped differently, and this is to help give an idea of the type of mapping we expect:
- Example 1 from https://swisscollnet.scnat.ch on an entomological collection
- Example 2 from https://swisscollnet.scnat.ch on a mineralogy collection
- Example 3 from http://rnc.humboldt.org.co on a botanical garden collection
- Example 4 from http://rnc.humboldt.org.co on aherbarium collection
NB: The Latimer core term objectClassificationName is very convenient to describe subsets of collections that do not necessarily have other ways of being grouped. For example, this is helpful for groups of non-monophyletic taxa (for example Algae). Ideally, the names used in this field should follow a controlled vocabulary. We haven’t set up one as we write though. If you need some guidance, I suggest following the DiSSCo discipline vocabulary for collections (see also DOI 10.3897/rio.10.e118244).
How to upload descriptors
Right now, only editors can upload descriptors to collections. If you are not a GRSciColl editor for the collection, institution or country of the collection and you would like to become one, please contact us at scientific-collections@gbif.org. You will need a GBIF account associated with your institutional email address.
Once you have your table of descriptors ready, the process to upload them is relatively straightforward.
- Go to the collection page that you would like to work on from https://scientific-collections.gbif.org/collection/search
- Click on the edit button (you will be redirected to the editing interface)
- Click on the “Descriptor group” tab (see the screenshot below)
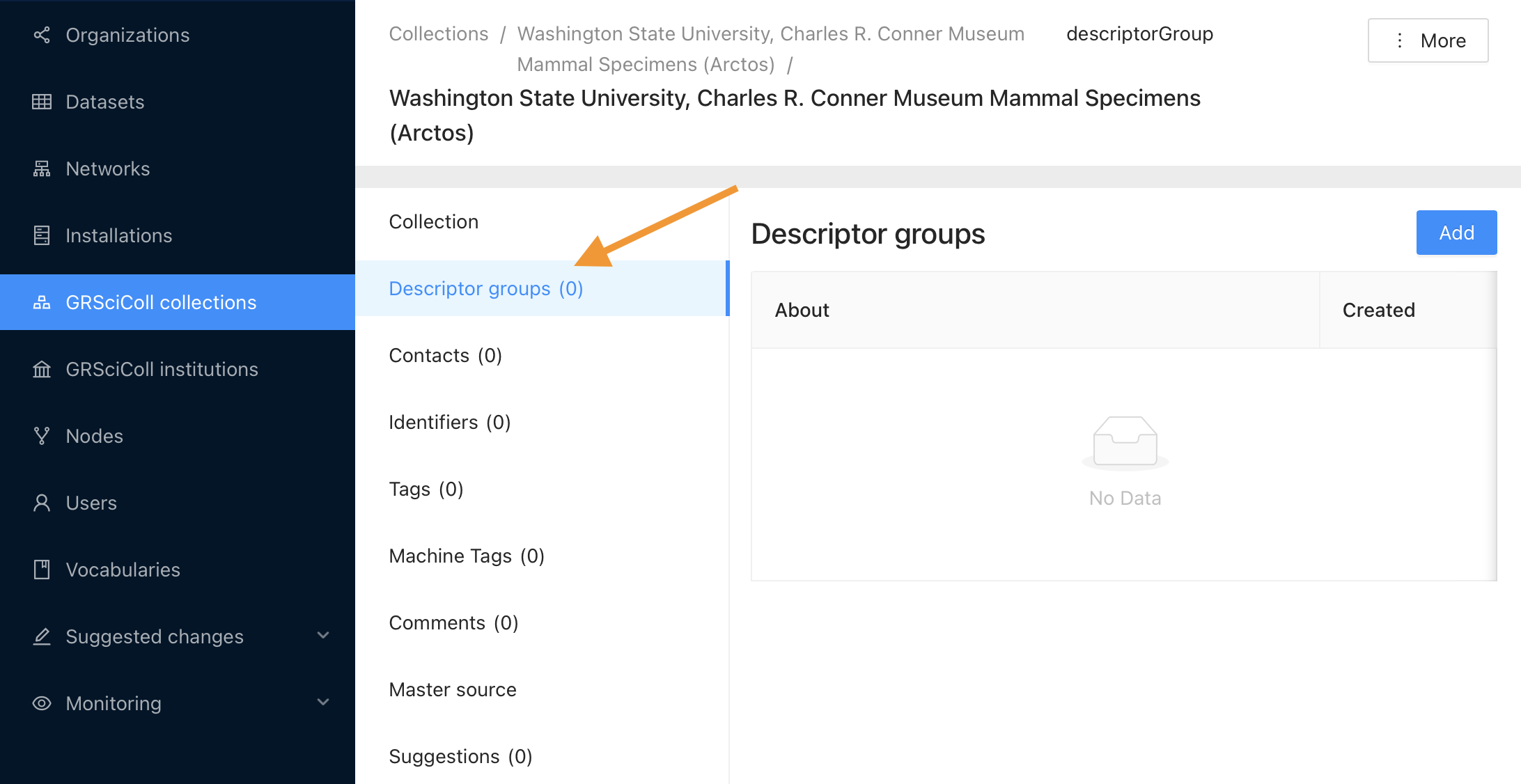
- Click Add and fill in the title and description of the descriptor group and choose your CSV file.
- Then click “add”.
Note that the API can also be used to upload collection descriptors programmatically. It is documented here. Please contact us at scientific-collections@gbif.org if you have any question.
Updating descriptors
You can update collection descriptors by clicking on the “edit” button of an existing descriptor group and replacing the table with a new one.
Improving collection descriptors and how to contribute
The collection descriptors are very new on GRSciColl and we would like to make them as useful to the community as possible. If you would like to give your input, please consider joining our mailing list or write a post on the GRSciColl section of our forum or leave us a comment here.
Here are a few points where we would like your input:
- Some descriptors would benefit from controlled vocabularies (like the ltc:objectClassificationName). Do you have suggestions or recommendations of existing vocabularies that we could use? Or specific values that you would like to see?
- We would like to index more descriptors as they are made available on GRSciColl. What type of descriptors would you like to share and see indexed?
- If you have unmappable data and would like to see them indexed in GRSciColl, please consider reaching out to the Latimer Core maintenance group or open a ticket in their GitHub repository.
Thank you very much! Happy collection describing.
