GBIF and Apache-Spark on Microsoft Azure tutorial
GBIF now has a snapshot of 1.3 billion occurrences✝ records on Microsoft Azure.
It is hosted by the Microsoft AI for Earth program, which hosts geospatial datasets that are important to environmental sustainability and Earth science. Hosting is convenient because you could now use occurrences in combination with other environmental layers and not need to upload any of it to the Azure. You can read previous discussions about GBIF and cloud computing here. The main reason you would want to use cloud computing is to run big data queries that are slow or impractical on a local machine.
In this tutorial, I will be running a simple query on 1.3 billion occurrences records. I will be using apache-spark. Spark has APIs in R, scala, and python.
✝The snapshot includes all records shared under CC0 and CC BY designations published through GBIF that have coordinates which have passed automated quality checks. The GBIF mediated occurrence data are stored in Parquet files in Azure Blob Storage in the West Europe Azure region. The periodic occurrence snapshots are stored in occurrence/YYYY-MM-DD, where YYYY-MM-DD corresponds to the date of the snapshot.
Setup for running a databricks spark notebook on Azure
You will be able to run free queries for a time, but eventually you will have to pay for your compute time.
Create a Microsoft Azure account here
I find it easiest to use the az command line interface. You can also set up using the Azure web interface. This tutorial is based off of the Databricks quickstart guide.
Download az here.
Log into your account and install databricks.
az login
az extension add --name databricks
Create a resource group gbif-resource-group. The GBIF snapshot is located in westeurope.
az group create --name gbif-resource-group --location westeurope
az account list-locations -o table
Create a workspacegbif-ws.
az databricks workspace create --resource-group gbif-resource-group --name gbif-ws --location westeurope --sku standard
From this page Azure web portal, click on gbif-resource-group.
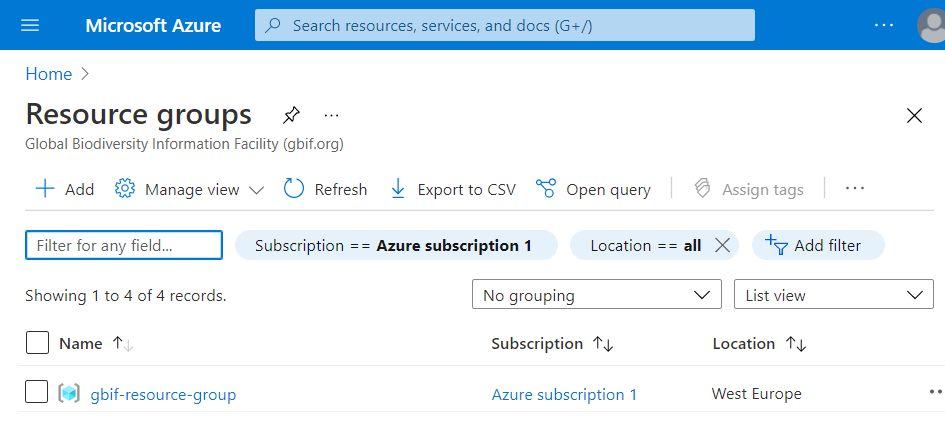
Click on launch workspace.
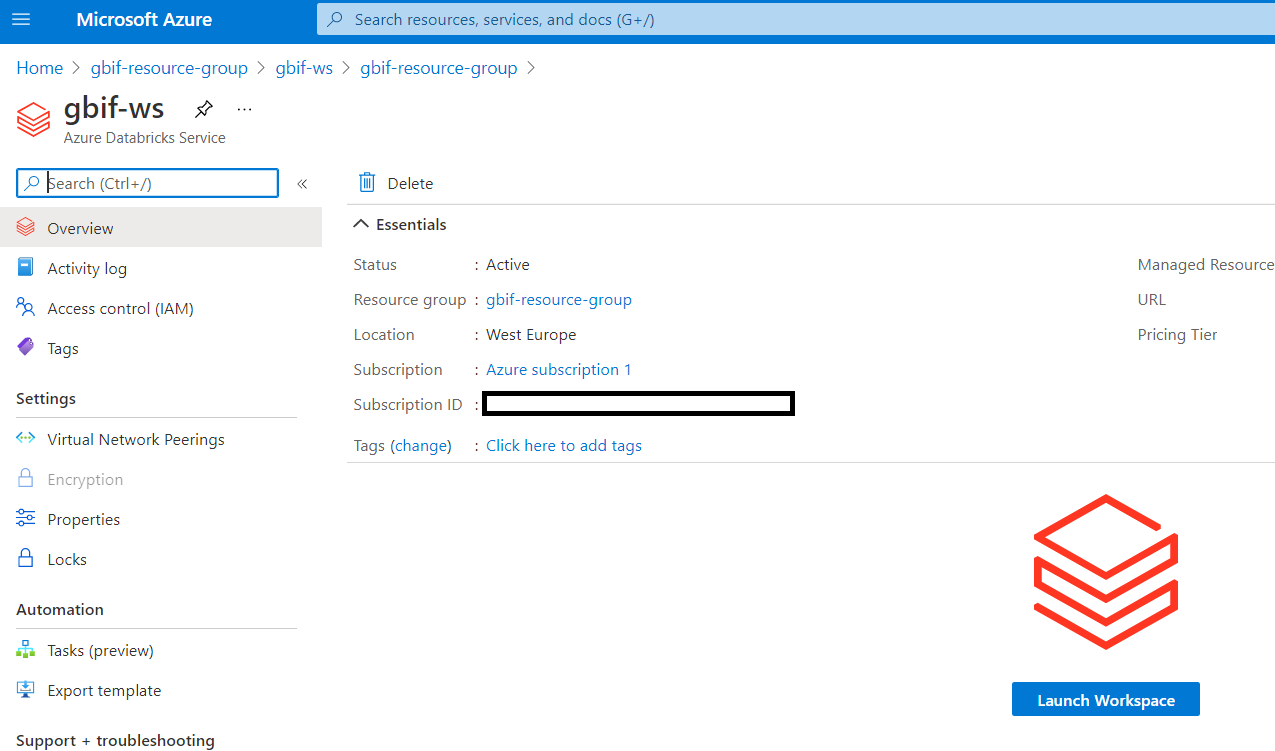
Click on new cluster.
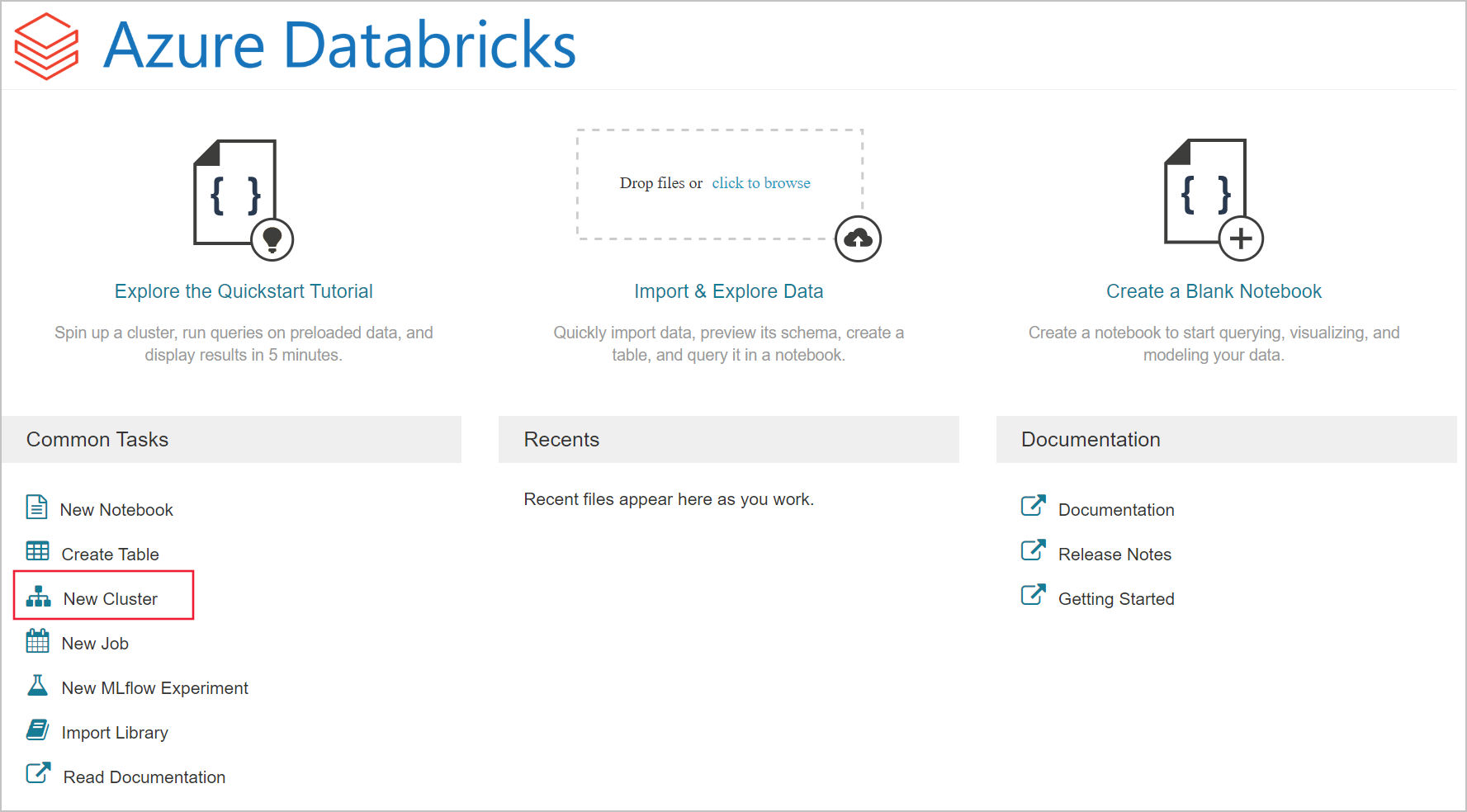
Create a new cluster named mysparkcluster. You can keep all of the default settings.
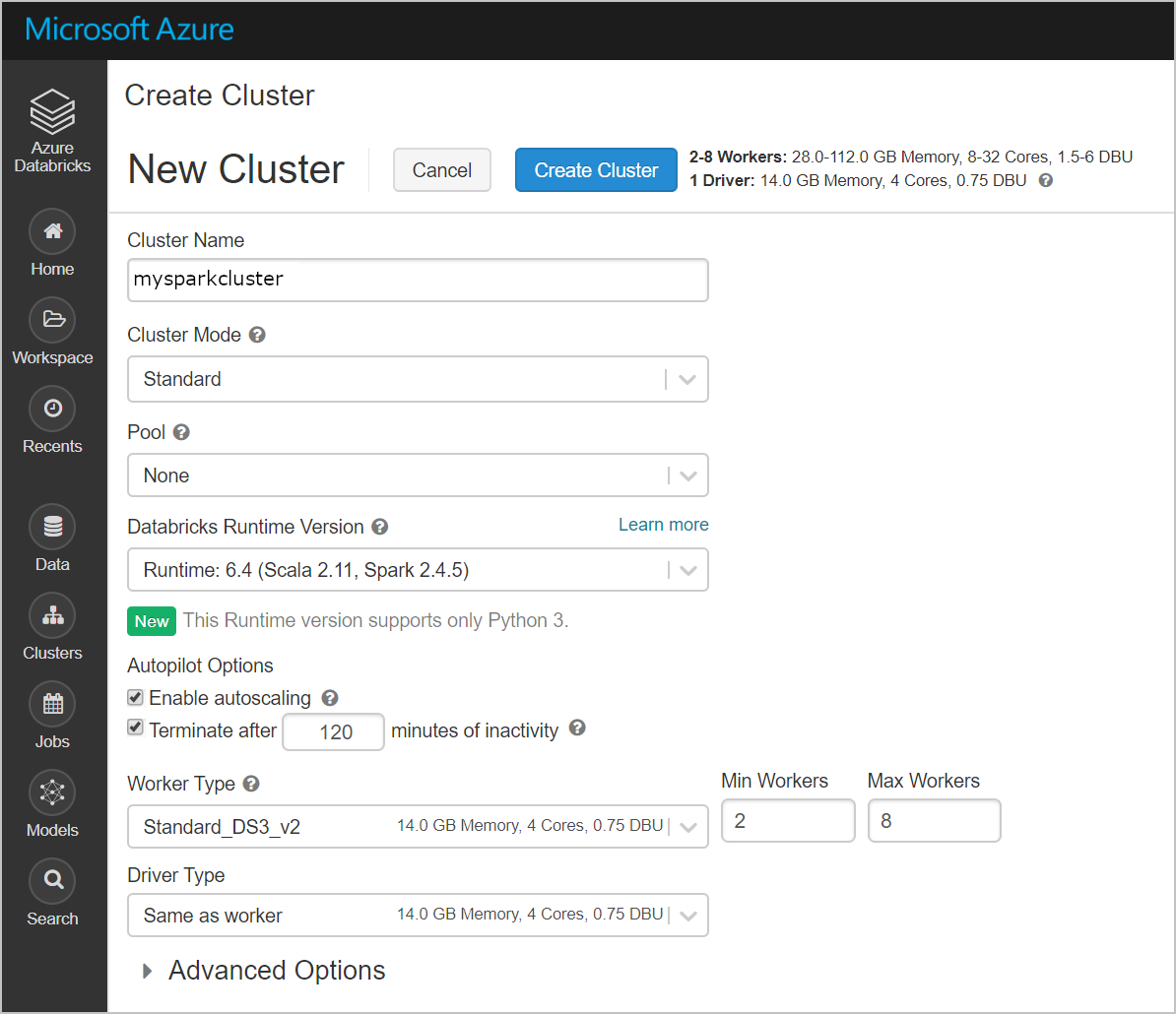
Click create blank notebook.
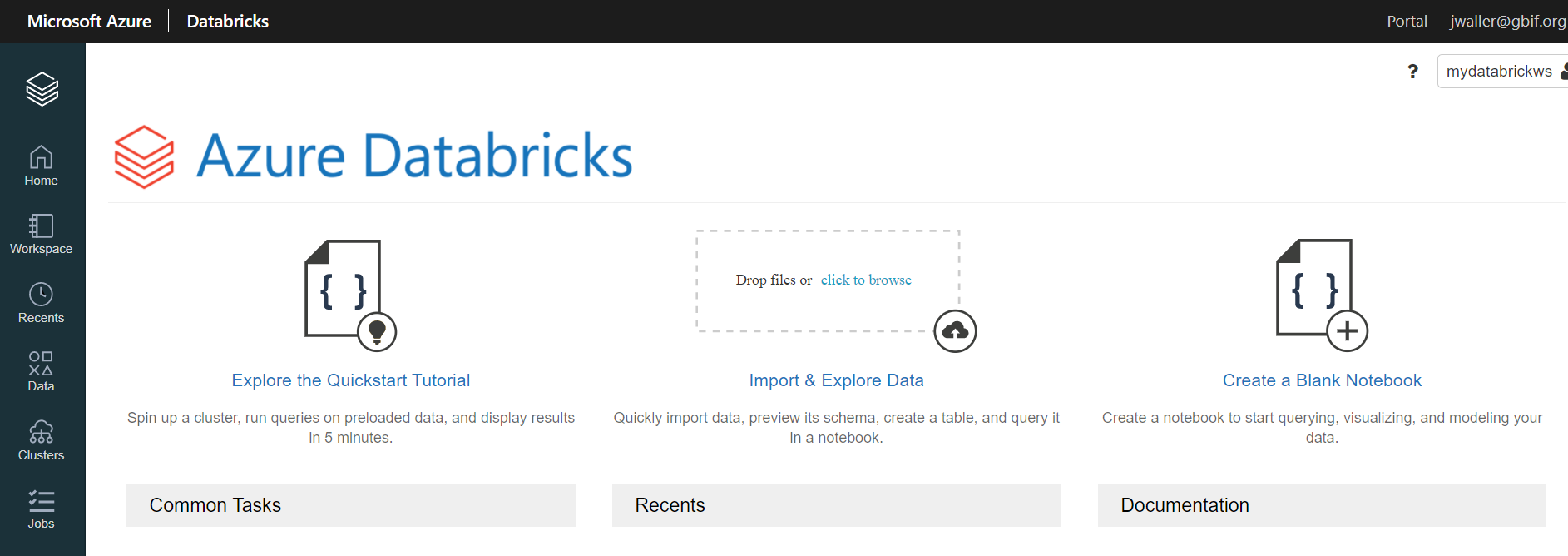
Select the default language you want to use (R, scala, python).
You should now be able to run the code below in your new notebook.
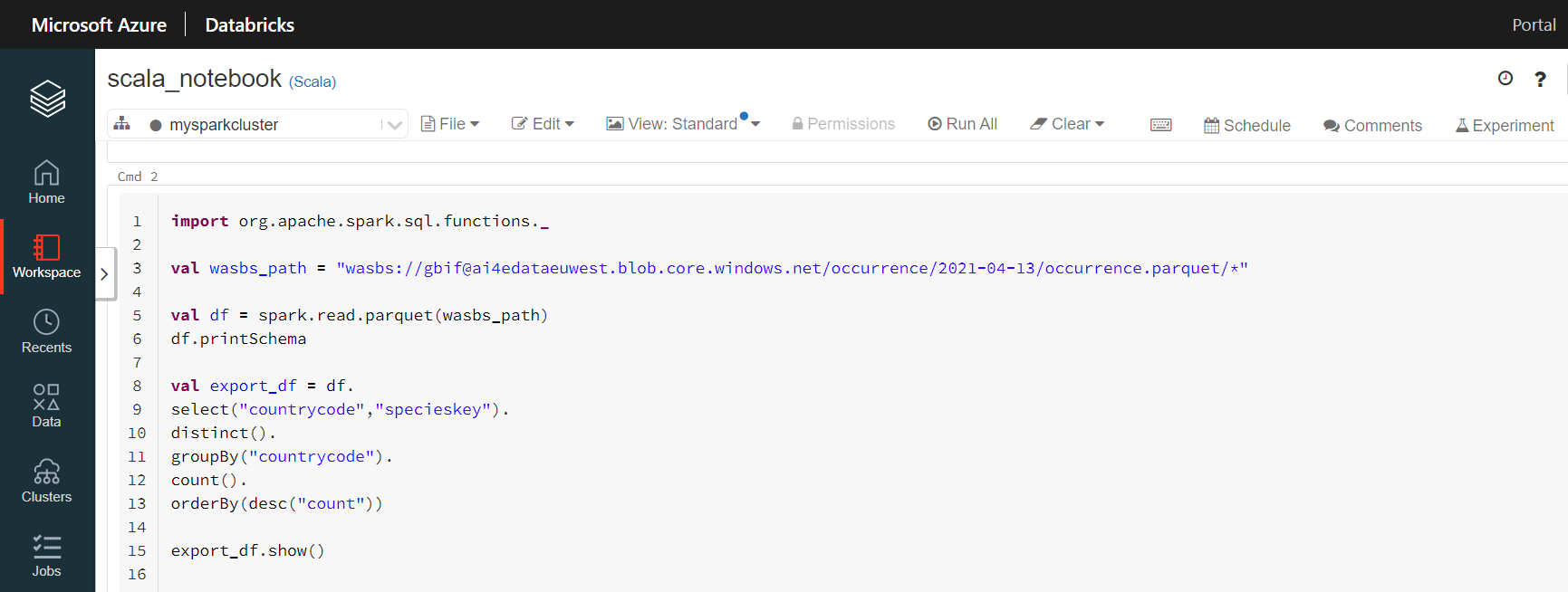
This code chunk will count the number of species (specieskeys) with occurrences in each country.
import org.apache.spark.sql.functions._
val gbif_snapshot_path = "wasbs://gbif@ai4edataeuwest.blob.core.windows.net/occurrence/2021-04-13/occurrence.parquet/*"
val df = spark.read.parquet(gbif_snapshot_path)
df.printSchema // show columns
df.count() // count the number of occurrences. 1.2B
// find the country with the most species
val export_df = df.
select("countrycode","specieskey").
distinct().
groupBy("countrycode").
count().
orderBy(desc("count"))
export_df.show()
The resulting export_df should look like this.
+-----------+------+
|countrycode| count|
+-----------+------+
| US|187515|
| AU|122746|
| MX| 86399|
| BR| 69167|
| CA| 64422|
| ZA| 60682|
Here is how you would count species by country using SparkR.
library(magrittr) # for %>%
install.packages("/databricks/spark/R/pkg", repos = NULL)
library(SparkR)
sc = sparkR.init()
sqlContext = sparkRSQL.init(sc)
gbif_snapshot_path = "wasbs://gbif@ai4edataeuwest.blob.core.windows.net/occurrence/2021-04-13/occurrence.parquet/*"
df = read.parquet(sqlContext,path=gbif_snapshot_path)
printSchema(df)
export_df = df %>%
select("countrycode","specieskey") %>%
distinct() %>%
groupBy("countrycode") %>%
count() %>%
arrange("count",decreasing = TRUE)
export_df %>% showDF()
How you would count species by country in PySpark.
from pyspark.sql import SQLContext
from pyspark.sql.functions import *
sqlContext = SQLContext(sc)
gbif_snapshot_path = "wasbs://gbif@ai4edataeuwest.blob.core.windows.net/occurrence/2021-04-13/occurrence.parquet/*"
# to read parquet file
df = sqlContext.read.parquet(gbif_snapshot_path)
df.printSchema # show columns
# find the country with the most species
export_df = df\
.select("countrycode","specieskey")\
.distinct()\
.groupBy("countrycode")\
.count()\
.orderBy(col("count").desc())
export_df.show()
Exporting a csv table
We would now like to export export_df from the previous section into a csv file, so we can analyze it on our own computer. There is also a bit of setup involved here.
It is easiest to use the command line tool az ( download here ) to set up storage accounts and containers for storing your exported csv.
Replace jwaller@gbif.org with your Microsoft-Azure account name.
az login
az storage account create -n gbifblobstorage -g gbif-resource-group -l westeurope --sku Standard_LRS
az role assignment create --role "Owner" --assignee "jwaller@gbif.org"
az role assignment create --role "Storage Blob Data Contributor" --assignee "jwaller@gbif.org"
az role assignment create --role "Storage Queue Data Contributor" --assignee "jwaller@gbif.org"
az storage container create -n container1 --account-name gbifblobstorage --auth-mode login
Run this command to get the secret key (sas_key) you will need in the next section.
az storage account keys list -g gbif-resource-group -n gbifblobstorage
The secret key (sas_key in next section) you want is under value.
{
"keyName": "key1",
"permissions": "FULL",
"value": "copy_me_big_long_secret_key_kfaldkfalskdfj203932049230492f_fakekey_j030303fjdasfndsafldkj=="
}
Now that the set up is over, you can export a dataframe as a csv file. The save path has the following form:
wasbs://container_name@storage_name.blob.core.windows.net/file_name.csv
We will use container1 and gbifblobstorage created earlier. Copy your sas_key to replace my fake one. Run this in one of the notebooks you set up earlier.
val sas_key = "copy_me_big_long_secret_key_kfaldkfalskdfj203932049230492f_fakekey_j030303fjdasfndsafldkj==" // fill the secret key from the previous section
spark.conf.set("fs.azure.account.key.gbifblobstorage.blob.core.windows.net",sas_key)
// or you could export a parquet file
// export_df.write.parquet("wasbs://container1@gbifblobstorage.blob.core.windows.net/export_df.parquet")
export_df.
coalesce(1).
write.
mode("overwrite").
option("header", "true").
option("sep", "\t").
format("csv").
save("wasbs://container1@gbifblobstorage.blob.core.windows.net/export_df.csv")
library(SparkR)
sparkR.session()
sas_key = "copy_me_big_long_secret_key_kfaldkfalskdfj203932049230492f_fakekey_j030303fjdasfndsafldkj=="
sparkR.session(sparkConfig = list(fs.azure.account.key.gbifblobstorage.blob.core.windows.net = sas_key))
export_df %>%
repartition(1) %>%
write.df("wasbs://container1@gbifblobstorage.blob.core.windows.net/export_df.csv", "csv", "overwrite")
sas_key = "copy_me_big_long_secret_key_kfaldkfalskdfj203932049230492f_fakekey_j030303fjdasfndsafldkj==" # fill the secret key from the previous section
spark.conf.set("fs.azure.account.key.gbifblobstorage.blob.core.windows.net",sas_key)
export_df\
.coalesce(1)\
.write\
.mode("overwrite")\
.option("header", "true")\
.option("sep", "\t")\
.format("csv")\
.save("wasbs://container1@gbifblobstorage.blob.core.windows.net/export_df.csv")
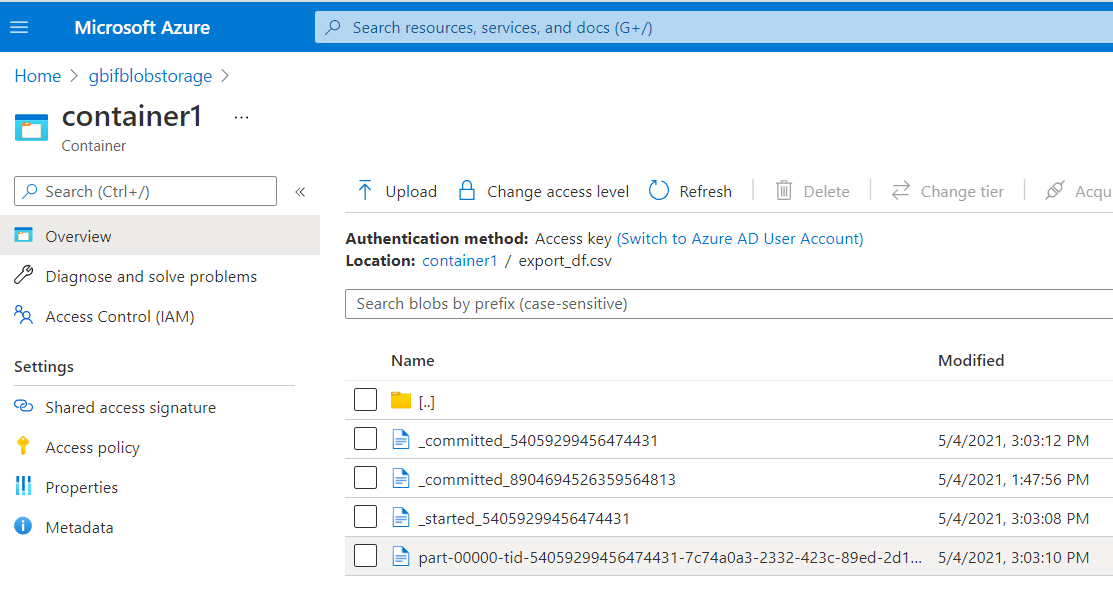
You can now download the export_df.csv from the azure web interface. The file will not be a single csv file but a directory named export_df. Within this directory you will find several files. The one you are interested in will be something looking like this:
export_df.csv/part-00000-tid-54059299456474431-7c74a0a3-2332-423c-89ed-2d1a98427a01-449-1-c000.csv
Downloading the csv
Go to your storage account.
Click on gbifblobstorage.

Click on containters.
Download the csv.
export_df.csv/part-00000-tid-54059299456474431-7c74a0a3-2332-423c-89ed-2d1a98427a01-449-1-c000.csv.
Citing custom filtered/processed data
If you end up using your exported csv file in a research paper, you will want a doi. GBIF now has a service for generating a citable doi from a list of involved datasetkeys with occurrences counts. See the GBIF citation guidelines and previous blog post.
You can generate a citation file for your custom dataset above using the following code chunk. Since our export_df.csv used all of the occurrences, we can simply group by datasetkey and count all of the occurrences to generate our citation.csv file.
val sas_key = "copy_me_big_long_secret_key_kfaldkfalskdfj203932049230492f_fakekey_j030303fjdasfndsafldkj==" // fill the secret key from the previous section
spark.conf.set("fs.azure.account.key.gbifblobstorage.blob.core.windows.net",sas_key)
import org.apache.spark.sql.functions._
val gbif_snapshot_path = "wasbs://gbif@ai4edataeuwest.blob.core.windows.net/occurrence/2021-04-13/occurrence.parquet/*"
val citation_df = spark.read.parquet(gbif_snapshot_path).
groupBy("datasetkey").
count()
citation_df.
coalesce(1).
write.
mode("overwrite").
option("header", "true").
option("sep", "\t").
format("csv").
save("wasbs://container1@gbifblobstorage.blob.core.windows.net/citation.csv")
You can also now use your citation file with the development version of rgbif to create a derived dataset and a citable DOI, although you would need to upload your exported dataset to Zenodo (or something similar) first.
# pak::pkg_install("ropensci/rgbif") # requires development version of rgbif
library(rgbif)
citation_data = readr::read_tsv("citation.csv")
# use derived_dataset_prep() if you just want to test
derived_dataset(
citation_data = citation_data,
title = "Research dataset derived from GBIF snapshot on MS Azure.",
description = "I used Azure and apache-spark to filter GBIF snapshot 2021-04-13.",
source_url = "https://zenodo.org/fake_upload",
user="your_gbif_user_name",
pwd="your_gbif_password"
)
The derived dataset with a citable DOI will appear on your gbif user page.
Hopefully you have everything that you need to start using GBIF mediated occurrence data on Microsoft Azure. Please leave a comment if something does not work for you.
